Metadata and parameters
Last updated on 2025-10-29 | Edit this page
Estimated time: 45 minutes
Overview
Questions
- How do I specify and configure parameters my whole workflow relies on?
- How do I set up parameters for individual jobs?
Objectives
- Be able to use the
configobject to accept external configuration - Be able to adjust the
configobject by using YAML files, and override individual options - Know how to format a metadata CSV file
- Be able to read a metadata CSV file and make use of it in a workflow
Global parameters
Thus far, each of our rules has taken one or more input files, and given output files solely based on that. However, in some cases we may want to control options without having them within an input file.
For example, in the previous episode, we wrote a rule to plot a graph
using the script src/plot_plaquette.py. The style of output
we got was good for a paper, but if we were producing a poster, or
putting the plot onto a slide with a dark background, we may wish to use
a different output style. The plot_plaquette.py script
accepts a --styles argument, to tell it what style file to
use to plot. One way to make use of this would be to add
--styles styles/paper.mplstyle directly to the
shell: block. However, if we had many such rules, and
wanted to switch from generating output for a paper to generating it for
a poster, then we would need to change the value in many places.
Instead, we can define a configuration file for our workflow. Create
a new file at config/config.yaml, defining the path to the
style file that we want:
Now, we can specify in our Snakefile to read this file by adding the following line near the top of the file:
configfile: "config/config.yaml"Then, when we use a script to generate a plot, we can update the
shell: block of the corresponding rule similarly to
"python src/plot_plaquette.py {input} --output_filename {output} --plot_styles {config[plot_styles]}"Snakemake will substitute the value it reads from the configuration
file in place of the {config[plot_styles]} placeholder.
(Note that unlike standard Python syntax, we don’t need quotes around
the plot_styles key string.)
Let’s double-check that our workflow still works, running
snakemake --cores 1 --forceall --printshellcmds --use-conda assets/plots/plaquette_scan.pdfNow that we have separated out our configuration from the workflow
itself, we can alter the configuration. For example, we may decide to
test plotting in the style of a different publication. We can test this
by changing jhep to prd, and running
snakemake --cores 1 --forceall --printshellcmds --use-conda assets/plots/plaquette_scan.pdfWe can see that the generated file now uses a different set of fonts.
Before continuing, let’s reset the workflow back to using the
jhep style.
Wilson flow
The tool su2pg_analysis.w0 computes the scale \(w_0\) given a log of the energy density
during evolution of the Wilson flow for an ensemble. To do this, the
reference scale \(\mathcal{W}_0\) needs
to be passed to the --W0 parameter. Use this, and the logs
stored in the files out_wflow for each ensemble’s raw data
directory, to output the \(w_0\) scale
in a file wflow.w0.json for each ensemble, taking the
reference value \(\mathcal{W_0} =
0.2\).
In config.yaml, add:
Then, add the following to the Snakefile:
# Compute w0 scale for single ensemble for fixed reference scale
rule w0:
input: "raw_data/{subdir}/out_wflow"
output: "intermediary_data/{subdir}/wflow.w0.json.gz"
conda: "envs/analysis.yml"
shell:
"python -m su2pg_analysis.w0 {input} --W0 {config[W0_reference]} --output_file {output}"Test this with:
$ snakemake --cores 1 --forceall --printshellcmds --use-conda intermediary_data/beta2.0/wflow.w0.json.gz
$ cat intermediary_data/beta2.0/wflow.w0.json.gz | gunzip | head -n 26
{
"program": "pyerrors 2.14.0",
"version": "1.1",
"who": "ed",
"date": "2025-10-08 20:28:56 +0100",
"host": "azusa, Linux-6.8.0-85-generic-x86_64-with-glibc2.39",
"description": {
"INFO": "This JSON file contains a python dictionary that has been parsed to a list of structures. OBSDICT contains the dictionary, where Obs or other structures have been replaced by DICTOBS[0-9]+. The field description contains the additional description of this JSON file. This file may be parsed to a dict with the pyerrors routine load_json_dict.",
"OBSDICT": {
"plaquette_w0": "DICTOBS0",
"clover_w0": "DICTOBS1"
},
"description": {
"group_family": "SU",
"num_colors": 2,
"nt": 48,
"nx": 24,
"ny": 24,
"nz": 24,
"beta": 2.0
}
},
"obsdata": [{
"type": "Obs",
"layout": "1",
"value": [2.2332066807096895],Generating different filetypes
In addition to different plot styles, we may also wish to generate different filetypes. PDF is useful for including in LaTeX, but SVG may be a better format to use with some tools.
Add a definition to your config.yaml file:
and update the output: block of the
plot_avg_plaquette rule as:
output:
multiext("assets/plots/plaquette_scan", config["plot_filetype"]),Note that unlike in the shell block, we can’t substitute
from config into an output string, so instead
we need to use the multiext helper function to append the
extension. (We could instead have used an f-string,
expand(), or any number of other ways to make this
substitution.)
Changing configurations
Now that we have separated out the data controlling our workflow from the code implementing it, we can individual overwrite parameters, or swap in an entirely different configuration file.
-
We would like to test plotting for a different journal. Re-run the workflow for the plaquette scan, using the
prd.mplstylestyle file, assnakemake --cores 1 --forceall --printshellcmds --use-conda --config plot_styles=styles/prd.mplstyle -- assets/plots/plaquette_scan.pdf(Note that we now need to add an extra
--to tell Snakemake that the list of--configoptions is complete.) We would like to create a plot to include in a poster, in SVG format. Create a new configuration file specifying to use the
poster.mplstylestyle file and the SVG file format, and re-run the workflow using the--configfileoption to specify this new file.
You should now have a file called something like
config/poster.yaml, with content
To test this, run
snakemake --cores 1 --forceall --printshellcmds --use-conda --configfile config/poster.yaml -- assets/plots/plaquette_scan.pdfSimilarly to the previous examples, we need to use the
-- option to tell Snakemake to stop trying to read more
config filenames.
Metadata from a file
We would frequently like our rules to depend on data that are specific to the specific ensembles being analysed. For example, consider the rule:
# Compute pseudoscalar mass and amplitude with fixed plateau
rule ps_mass:
input: "raw_data/{subdir}/out_corr"
output: "intermediary_data/{subdir}/corr.ps_mass.json.gz"
conda: "envs/analysis.yml"
shell:
"python -m su2pg_analysis.meson_mass {input} --output_file {output} --plateau_start 18 --plateau_end 23"This rule hardcodes the positions of the start and end of the plateau region. In most studies, each ensemble and observable may have a different plateau position, so there is no good value to hardcode this to. Instead, we’d like a way of picking the right value from some list of parameters that we specify.
We could do this within the Snakefile, but where possible it is good to avoid mixing data with code. We shouldn’t need to modify our code every time we add or modify the data it is analysing. Instead, we’d like to have a dedicated file containing these parameters, and to be able to have Snakemake read it and pick out the correct values.
To do this, we can exploit the fact that Snakemake is an extension of Python. In particular, Snakemake makes use of the Pandas library for tabular data, which we can use to read in a CSV files.
CSVs aren’t the only way to do this; for more complex data, YAML or even JSON may be a better choice. But CSV is good for most purposes, and easier to get started with. It’s also more readable for non-specialists investigating the workflow, which is valuable in and of itself.
Let’s add the following to the top of the file:
import pandas
metadata = pandas.read_csv("metadata/ensemble_metadata.csv")The file being read here is a CSV (Comma Separated Values) file. We can create, view, and modify this with the spreadsheet tool of our choice. Let’s take a look at the file now.
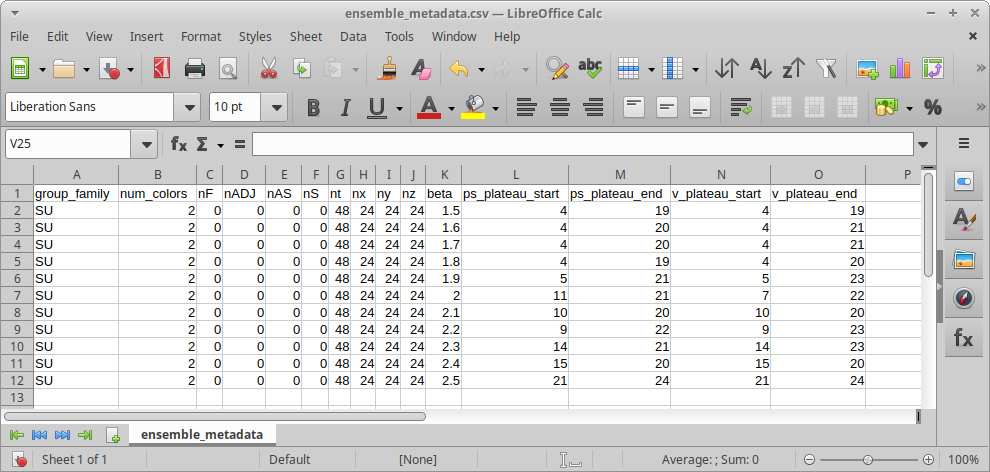
You can see that we have columns defining metadata to identify each ensemble, and columns for parameters relating to the analysis of each ensemble.
Now, how do we tell Snakemake to pull out the correct value from this?
# Compute pseudoscalar mass and amplitude, read plateau from metadata
rule ps_mass:
input: "raw_data/beta{beta}/out_corr"
output: "intermediary_data/beta{beta}/corr.ps_mass.json.gz"
params:
plateau_start=lookup(within=metadata, query="beta == {beta}", cols="ps_plateau_start"),
plateau_end=lookup(within=metadata, query="beta == {beta}", cols="ps_plateau_end"),
conda: "envs/analysis.yml"
shell:
"python -m su2pg_analysis.meson_mass {input} --output_file {output} --plateau_start {params.plateau_start} --plateau_end {params.plateau_end}"We’ve done a couple of things here. Firstly, we’ve made explicit the
reference to \(\beta\) in the file
paths, so that we can use beta as a wildcard, similarly to
in the challenge in the previous episode. Secondly, we’ve introduced a
params: block. This is how we tell Snakemake about
quantities that may vary from run to run, but that are not filenames.
Thirdly, we’ve used the lookup() function to search the
metadata dataframe for the ensemble that we are
considering. Finally, we’ve used {params.plateau_start} and
{params.plateau_end} placeholders to use these parameters
in the shell command that gets run.
Let’s test this now:
$ snakemake --cores 1 --forceall --printshellcmds --use-conda intermediary_data/beta2.0/corr.ps_mass.json.gz
$ cat intermediary_data/beta2.0/corr.ps_mass.json.gz | gunzip | head -n 29
{
"program": "pyerrors 2.14.0",
"version": "1.1",
"who": "ed",
"date": "2025-10-08 20:41:12 +0100",
"host": "azusa, Linux-6.8.0-85-generic-x86_64-with-glibc2.39",
"description": {
"INFO": "This JSON file contains a python dictionary that has been parsed to a list of structures. OBSDICT contains the dictionary, where Obs or other structures have been replaced by DICTOBS[0-9]+. The field description contains the additional description of this JSON file. This file may be parsed to a dict with the pyerrors routine load_json_dict.",
"OBSDICT": {
"mass": "DICTOBS0",
"amplitude": "DICTOBS1"
},
"description": {
"group_family": "SU",
"num_colors": 2,
"representation": "fun",
"nt": 48,
"nx": 24,
"ny": 24,
"nz": 24,
"beta": 2.0,
"mass": 0.0,
"channel": "ps"
}
},
"obsdata": [{
"type": "Obs",
"layout": "1",
"value": [2.1988677698535195],Vector mass
Add a rule to compute the vector meson mass and amplitude, using the
columns beginning v_ in the ensemble metadata file for the
plateau limits.
Hint: su2pg_analysis.meson_mass accepts an argument
--channel, which defaults to ps.
This is very close to the rule for the PS mass.
# Compute vector mass and amplitude, read plateau from metadata
rule v_mass:
input: "raw_data/beta{beta}/out_corr"
output: "intermediary_data/beta{beta}/corr.v_mass.json.gz"
params:
plateau_start=lookup(within=metadata, query="beta == {beta}", cols="v_plateau_start"),
plateau_end=lookup(within=metadata, query="beta == {beta}", cols="v_plateau_end"),
conda: "envs/analysis.yml"
shell:
"python -m su2pg_analysis.meson_mass {input} --channel v --output_file {output} --plateau_start {params.plateau_start} --plateau_end {params.plateau_end}"We can again verify this using
$ snakemake --cores 1 --forceall --printshellcmds --use-conda intermediary_data/beta2.0/corr.ps_mass.json.gz
$ cat intermediary_data/beta2.0/corr.ps_mass.json.gz | gunzip | head -n 29
{
"program": "pyerrors 2.14.0",
"version": "1.1",
"who": "ed",
"date": "2025-10-08 20:43:06 +0100",
"host": "azusa, Linux-6.8.0-85-generic-x86_64-with-glibc2.39",
"description": {
"INFO": "This JSON file contains a python dictionary that has been parsed to a list of structures. OBSDICT contains the dictionary, where Obs or other structures have been replaced by DICTOBS[0-9]+. The field description contains the additional description of this JSON file. This file may be parsed to a dict with the pyerrors routine load_json_dict.",
"OBSDICT": {
"mass": "DICTOBS0",
"amplitude": "DICTOBS1"
},
"description": {
"group_family": "SU",
"num_colors": 2,
"representation": "fun",
"nt": 48,
"nx": 24,
"ny": 24,
"nz": 24,
"beta": 2.0,
"mass": 0.0,
"channel": "v"
}
},
"obsdata": [{
"type": "Obs",
"layout": "1",
"value": [2.213089845075537],If it seems awkward to need to define multiple rules that differ only in which channel they look at, this is a good point, and one that we will deal with in the episode on Awkward Corners.
- Use a YAML file to define parameters to the workflow, and attach it
using
configfile:near the top of the file. - Override individual options at run-time with the
--configoption. - Load additional parameter files at run-time using the
--configfileoption. - Use a CSV file loaded into a Pandas dataframe to load ensemble-specific metadata.
- Use
lookup()to get information out of the dataframe in a rule. - Use
params:to define job-specific parameters that do not describe filenames.
