Publishing in open science repositories
Overview
Teaching: 20 min
Exercises: 20 minQuestions
How is an open science repository different to something like GitHub?
How do I create a permanent version of record of my code?
How can I create a DOI for my code and cite it?
Objectives
Understand the difference between the various types of repository.
Understand how to push a repository from GitHub to Zenodo.
Be able to find the DOI for a repository and cite it.
So far we have been working to tidy our repository and make it reproducible, pushing and pulling work to and from GitHub. However, at the end of any drafting process there comes a point where you declare the work finished, and make it available as a finalised version of record—for a paper this is where you submit to a journal and (hopefully) have the paper published. Similarly, we would like to make a finalised version of our code available, to give an indication that:
- The code is finalised, and the version presented is the version used to perform the analysis for any publications based on the code. By analogy, we wouldn’t link colleagues to the online collaborative editable version of our paper instead of publishing in a journal.
- The code will be available for the long term. While GitHub has been around for a while, it makes no promises as to long-term availability. Published academic journals are available from decades or centuries ago; drafts and collaborative notes from that period are harder to find.
- We would like the code to be treated (and cited) as an academic output. GitHub is popular with and well-known among software engineers, but in academic circles it is less familiar, or at least is known as a website to get free software from; the idea of software (code) being valuable in and of itself is more novel. Having a repository similar to the more familiar preprint repositories, and that can assign a DOI to a peice of work, will hopefully better align your work as an object worthy of academic consideration.
DOI?
DOI, short for Digital Object Identifier, is a type of unique identifier most frequently used to identify academic journal articles, but which can also be used for other “digital objects”, including data sets as well as, for example, official publications of the European Union. It is designed to be more permanent than a URL, since internet locations may change over time as journals or their publishers change names. They can look like
doi:NNNN.MM/XXXwhereNNNN,MM, andXXXare numbers; they can also be formatted ashttps://doi.org/NNNN.MM/XXX. The latter can be entered into a browser, anddoi.orgwill always redirect to the current location of the object.
Publish in a data repository
One good option for publishing code is to use a data repository. There is a wide range of data repositories available, many of which are specialist towards particular disciplines. Many potential venues for publishing software (and many other digital services to enable open science) can be searched at the European Open Science Cloud. It’s also possible that your institution hosts its own data repository, and/or mandates a specific one. Your institution’s Open Research or Open Data policy, if it has one, should have more detail on this.
Today we’ll look at a general-purpose data repository called Zenodo. Zenodo is hosted by CERN (the European Centre for Nuclear Research in Geneva), and is part of the same infrastructure that manages the collossal amounts of data generated by the Large Hadron Collider. The computing resources supporting it have operational plans and budgets reaching decades into the future, which is about as long-term as any services currently operate on.
Keep your tests separate from your real data
Because Zenodo is designed to keep research data safe for the long term, it’s not a good idea to upload test or toy data to it. For that reason, for this lesson we’ll be using the “sandbox” version of Zenodo. This behaves exactly like the real version, but is regularly purged of new datasets, and isn’t where you should publish your actual research outputs! Remember to go to the real Zenodo once you’re ready to publish your research.
To publish on Zenodo (and Zenodo Sandbox), we first need to create an account.
While this can be done manually, the quickest way for our purposes is to log in using GitHub. This will connect our GitHub account with our Zenodo account. If you have one, you can also link your ORCID to your Zenodo account, so that your publications on Zenodo are easier to tie back to your work in other venues.

Once we are signed up and logged into Zenodo (Sandbox), we can use the drop-down next to our email address at the top-right to select the GitHub option.
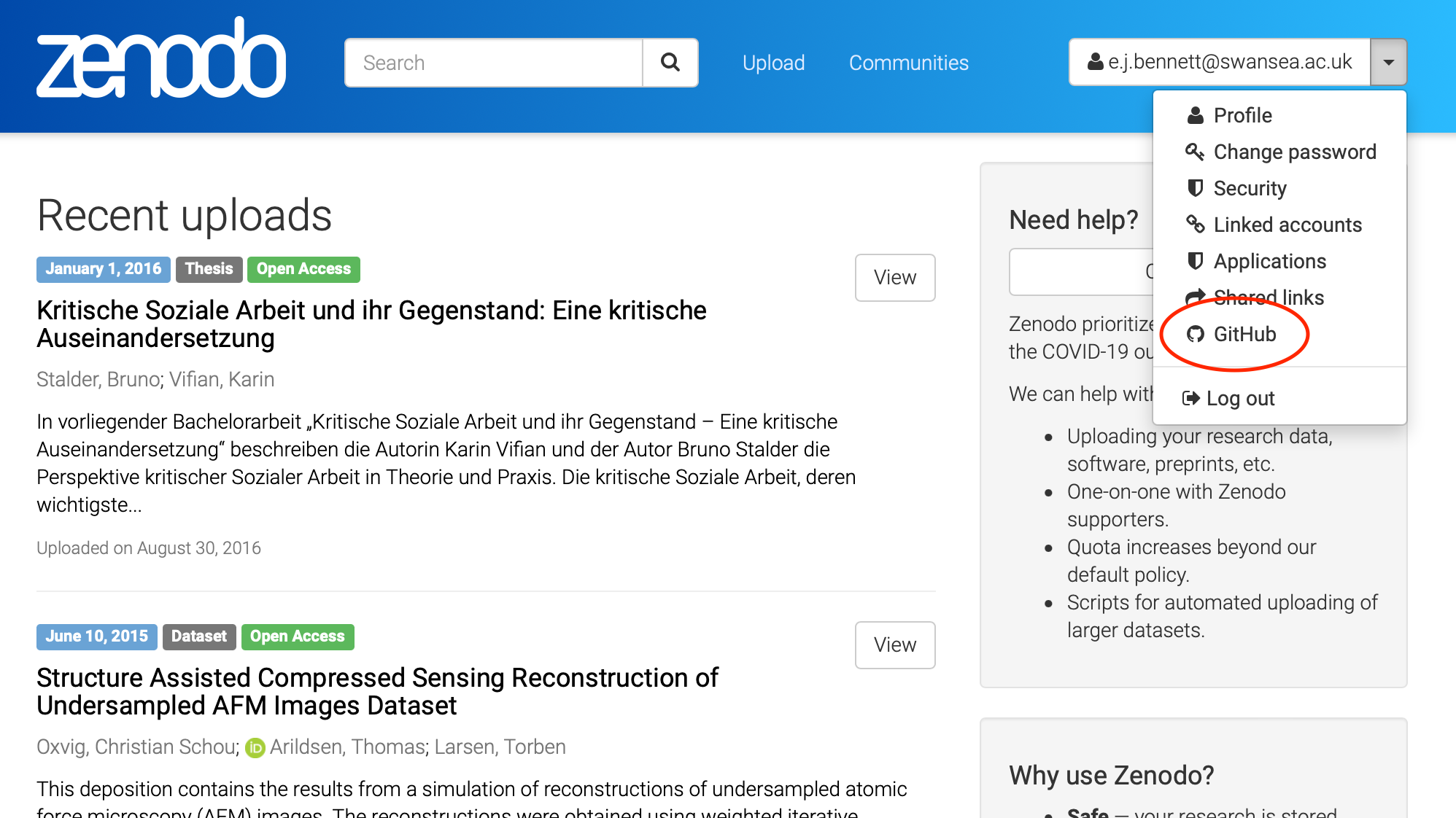
This gives us a
three-step process to publish our repository to Zenodo. Firstly, out of the list of
repositories presented (found from our GitHub account), we choose which we want to publish
to Zenodo. Let’s do that now for the zipf repository.
Once this is done, we need to create a “release” of our code on GitHub.
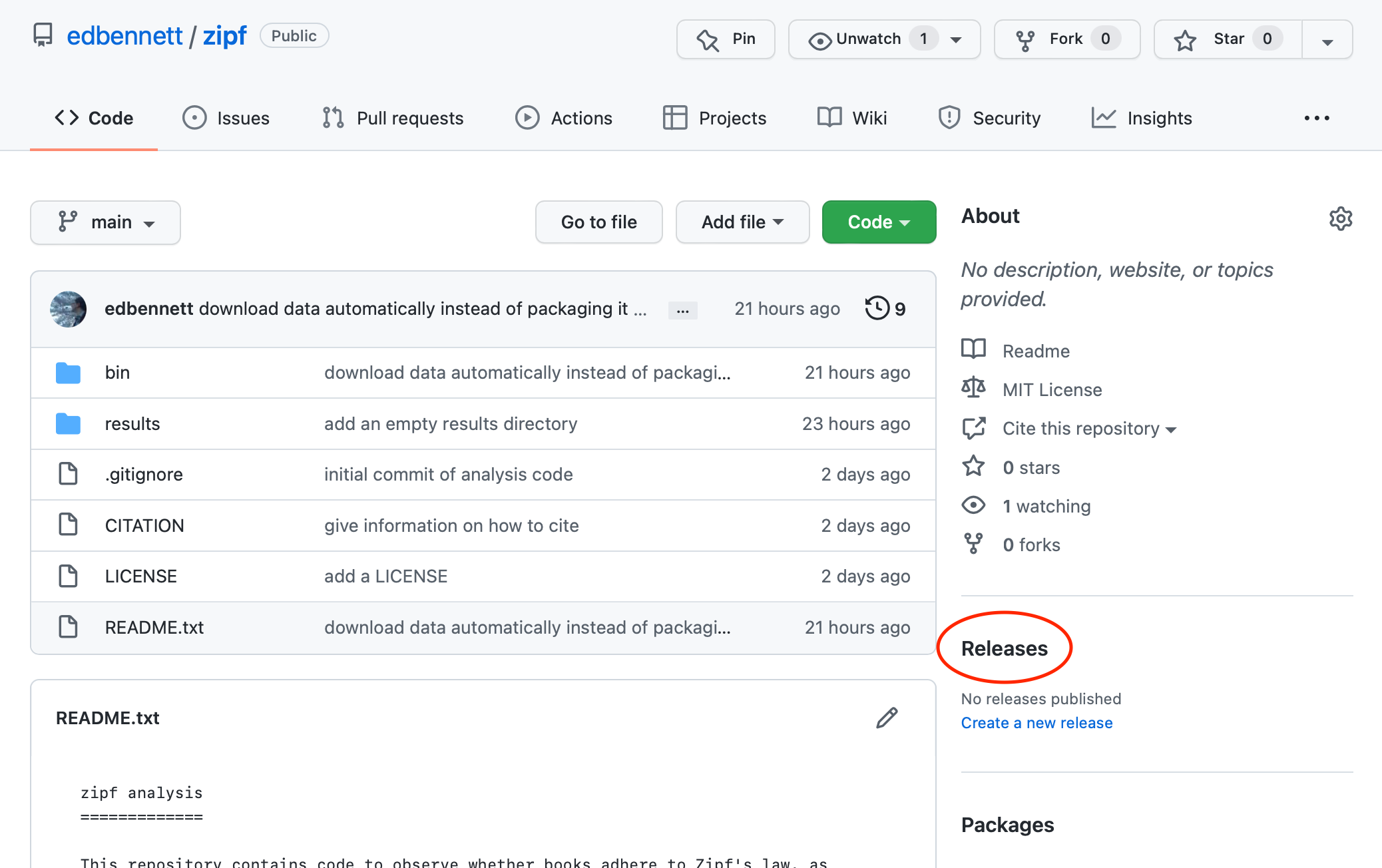
To do this, we can click through from the repository home page to the Releases page, and from there click the “Create a new release” button.
The form to create a new release requests some information
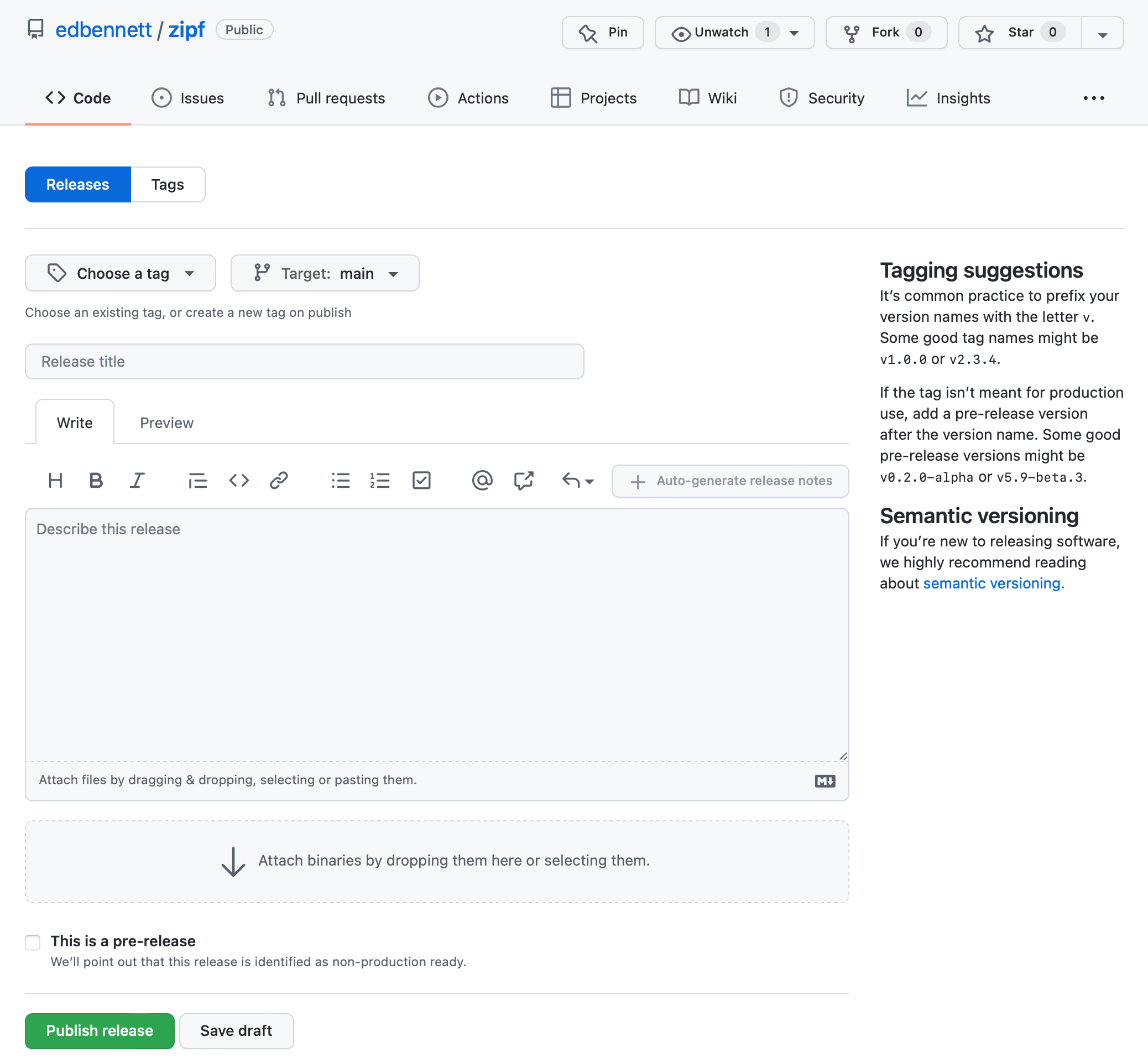
- Choose a tag: If you use tags in your repository, you can choose which tagged commit should be released. If not, then GitHub can automatically create a tag for you when you publish the release—type the name you want the tag to have in this box. This will frequently be the same as the release name.
- Target: this should be the branch that you are releasing from. This will most likely
be the
mainbranch. - Release title: This should be a version name for the particular release. This is
designed for software that gets regular releases, or at least more than one in its
lifetime. For a piece of data analysis code that is specific to one publication, it
is enough to use
v1.0.0here. - Describe the release: Some more discussion of the release can in principle go here. Note that this is describing the release, not the code—the code (and underlying research) should be discussed in the README. This is again more useful when you are releasing multiple times; each release should use the description to outline the changes included since the previous release.
- Attach binaries: We can ignore this; if we were publishing software written in a language like C or Fortran, and that we wanted others to be able to install, we could attach the compiled versions of the program here.
Once we have filled out the form, we can click Publish, and Zenodo (or the Sandbox) will pick up that we have created a release, and will create a corresponding data publication. It will also then create a DOI, which you can add to the bibliography of any papers that use results generated by the code. This will take a while (a few minutes to an hour); you can check the status by clicking on the repository in the listing on Zenodo.
The third step suggested by Zenodo is to add a badge to our README. Since we have used plain text for our README, that will not work, but if you were using Markdown, you could copy and paste the code generated by Zenodo to add the DOI of the repository, so that readers arriving to the GitHub repository page are aware that they can cite the code via DOI as well.
Alternatives publishing routes
For tools that can be applied more broadly, there are some alternative routes to publication (that can be used instead of or in addition to Zenodo).
- Publish a package. Depending on the venue, this may or may not aid citability. For example, publishing a Pip package will not enable citation, but publishing to WorkflowHub publicly will create a DOI.
- Publish a paper. There are publications that specifically enable promotion and constructive peer review of research software. The Journal of Open Source Software connects to GitHub similarly to Zenodo, but requires a very short “paper” explaining the context of what the software does. This allows you to get some feedback from other computational researchers on your code.
Home turf
Try and find your own institution’s policy on open research or open data. Does it have a local repository for source code (or research data more generally), or recommendations on where to publish it?
Fitting in
Are there any discipline-specific repositories for data or code that are in use in your discipline?
Discuss with a neighbour, or in breakout rooms.
Versioning
Wei published the analysis code for his last publication about bat biomechanics on Zenodo. Now he has prepared a new paper about squirrel biomechanics, making use of a lot of the same code, but with some modifications to make it applicable to squirrels rather than bats. He is wondering how best to publish this modified code. What would be the best option for Wei?
- Publish the new version of the code entirely separately, with a citation to the bat code.
- Publish the new version of the code entirely separately, with no mention of the bat code.
- Create a new version of the previous published code on Zenodo, and update the metadata to indicate that the code is now squirrel-specific.
- Keep the updated code on GitHub, but keep citing the DOI for the bat version of the code, since it links back to GitHub where the updated code is.
- Something else?
Publish the challenge
Enable preservation of the
challengerepository in the Zenodo Sandbox. Create a new release, and get the DOI from Zenodo.
What next?
In this lesson we have focused primarily on taking a piece of code that has already been written and adjusting it to prepare it for publication. Of course, as you do this, your habits in writing software will change so that for future publications the workload is less—perhaps you keep things in Git from the start, choose a directory structure, and write a README as you progress through your work. As you gain experience, the process of publishing your analysis code will become second-nature.
If you do continue down this road, you might find it useful to have tools that nudge you towards a neater structure from the moment you start your project. One such tool that is also popular with professional software engineers is called Cookiecutter; this gives you access to a variety of project templates that are appropriate for particular types of project. In particular, there is a Cookiecutter template for data science that will give you a project structure similar to the one we have discussed in this lesson (along with some other suggestions that we haven’t had time to cover).
Key Points
Source code hosts like GitHub are designed for active development of software. Open science repositories are to keep versions of record for the longer term.
Services like Zenodo allow you to package a particular commit, archive it, and give it a permanent identifier.
Many services like Zenodo will automatically give you a DOI for any dataset, including repositories pulled from GitHub.
DOIs for code repositories can be cited in journal articles the same way as any other publication.